D.A. Forsyth --- 3310 Siebel Center


TA's:
You should do this homework on your own -- one submission per student, and by submitting you are certifying the homework is your work.
Submission: Course submission policy is here
I strongly advise you use the R language for this homework (but word is out on Piazza that you could use Python; note I don't know if packages are available in Python). You will have a place to upload your code with the submission.
The UC Irvine machine learning data repository hosts a famous collection of data on whether a patient has diabetes (the Pima Indians dataset), originally owned by the National Institute of Diabetes and Digestive and Kidney Diseases and donated by Vincent Sigillito. You can find this data at http://archive.ics.uci.edu/ml/datasets/Pima+Indians+Diabetes. You should look over the site and check the description of the data. In the "Data Folder" directory, the primary file you need is named "pima-indians-diabetes.data". This data has a set of attributes of patients, and a categorical variable telling whether the patient is diabetic or not. For several attributes in this data set, a value of 0 may indicate a missing value of the variable.
You should use a normal distribution to model each of the class-conditional distributions. You should write this classifier yourself (it's quite straight-forward).
Report the accuracy of the classifier on the 20% evaluation data, where accuracy is the number of correct predictions as a fraction of total predictions.
Report the accuracy of the classifier on the 20% that was held out for evaluation.
train (features, labels, classifier, trControl=trainControl(method='cv',number=10))
The klaR package can estimate class-conditional densities using a density estimation procedure that I will describe much later in the course. I have not been able to persuade the combination of caret and klaR to handle missing values the way I'd like them to, but that may be ignorance (look at the na.action argument).Report the accuracy of the classifier on the held out 20%
svmlight (features, labels, pathsvm)
You don't need to understand much about SVM's to do this as we'll do that in following exercises. You should NOT substitute NA values for zeros for attributes 3, 4, 6, and 8.Using the predict function in R, report the accuracy of the classifier on the held out 20%
Hint If you are having trouble invoking svmlight from within R Studio, make sure your svmlight executable directory is added to your system path. Here are some instructions about editing your system path on various operating systems: https://www.java.com/en/download/help/path.xml You would need to restart R Studio (or possibly restart your computer) afterwards for the change to take effect.
For this assignment, you should do your coding in R once again, but you may use libraries for the algorithms themselves.
The MNIST dataset is a dataset of 60,000 training and 10,000 test examples of handwritten digits, originally constructed by Yann Lecun, Corinna Cortes, and Christopher J.C. Burges. It is very widely used to check simple methods. There are 10 classes in total ("0" to "9"). This dataset has been extensively studied, and there is a history of methods and feature construc- tions at https://en.wikipedia.org/wiki/MNIST_database and at the original site, http://yann.lecun.com/exdb/mnist/ . You should notice that the best methods perform extremely well.
There is also a version of the data that was used for a Kaggle competition. I used it for convenience so I wouldn't have to decompress Lecun's original format. I found it at http://www.kaggle.com/c/digit-recognizer .
If you use the original MNIST data files from http://yann.lecun.com/exdb/mnist/ , the dataset is stored in an unusual format, described in detail on the page. You should begin by reading over the technical details. Writing your own reader is pretty simple, but web search yields readers for standard packages. There is reader code for R available (at least) at https://stackoverflow.com/questions/21521571/how-to-read-mnist-database-in-r . Please note that if you follow the recommendations in the accepted answer there at https://stackoverflow.com/a/21524980 , you must also provide the readBin call with the flag signed=FALSE since the data values are stored as unsigned integers. You need to use R for this course, but for additional reference, there is reader code in MATLAB available at http://ufldl.stanford.edu/wiki/index.php/Using_the_MNIST_Dataset .
Regardless of which format you find the dataset stored in, the dataset consists of 28 x 28 images. These were originally binary images, but appear to be grey level images as a result of some anti-aliasing. I will ignore mid grey pixels (there aren't many of them) and call dark pixels "ink pixels", and light pixels "paper pixels"; you can modify the data values with a threshold to specify the distinction, as described here https://en.wikipedia.org/wiki/Thresholding_(image_processing) . The digit has been centered in the image by centering the center of gravity of the image pixels, but as mentioned on the original site, this is probably not ideal. Here are some options for re-centering the digits that I will refer to in the exercises.
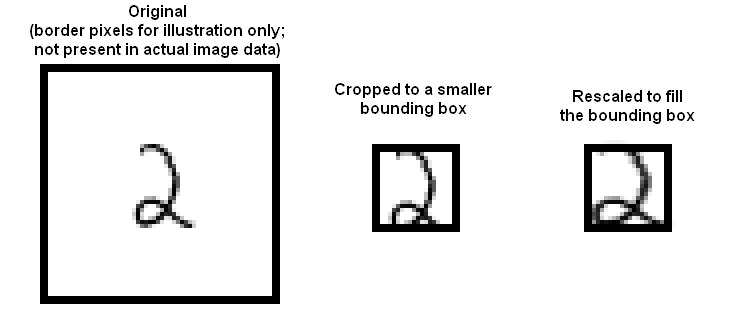
Investigate classifying MNIST using naive Bayes. Fill in the accuracy values for the four combinations of Gaussian v. Bernoulli distributions and untouched images v. stretched bounding boxes in a table like this. Please use 20 x 20 for your bounding box dimensions.
| Accuracy | Gaussian | Bernoulli |
|---|---|---|
| Untouched images | ||
| Stretched bounding box |
Which distribution (Gaussian or Bernoulli) is better for untouched pixels? Which is better for stretched bounding box images?
| Accuracy | depth = 4 | depth = 8 | depth = 16 |
|---|---|---|---|
| #trees = 10 | |||
| #trees = 20 | |||
| #trees = 30 |